Rubisco purification protocol (15Q)
Published:
Protocol for the high quality purification of Rubisco, applied to green algae, seaweed and plants, yielding active, intact Rubisco holoenzyme complexes. This protocol was developed from an original protocol that included sucrose gradient centrifugation, which was limiting for the throughput of the purification. In this protocol, a 15Q ion exchange column is used. Lower quality ion exchange resins (e.g. HiTrap Q XL) are not appropriate for this.
last updated: 24/07/25
Before starting
Materials
*Essential
- *Ammonium sulphate (e.g.; A4418)
- *DTT (dithiothreitol) (e.g.; R0861)
- *SOURCE 15Q ion exchange column/resin (e.g.; #17515901)
- *Superdex 200 gel filtration column (e.g.; #28989335)
- Protein concentration columns (e.g.; Amicon® Ultra-15)
Buffers
Rubisco lysis buffer
- 50 mM Tris-HCl
- 100 mM NaCl
- 0.5 mM EDTA (pH 8.0)
- 5 mM DTT (added fresh)
- Adjust to pH 8.0
IEX buffer (A)
- 50 mM Tris-HCl
- 50 mM NaCl
- 1 mM DTT
- Adjust to pH 8.0
IEX elution buffer (B)
- 50 mM Tris-HCl
- 1 M NaCl
- 1 mM DTT
- Adjust to pH 8.0
SEC Buffer
- 50 mM Tris-HCl
- 50 mM NaCl
- 5% glycerol (w/v)
- 1 mM DTT
- Adjust to pH 8.0
Essential note
Ensuring that the Rubisco remains in a reducing environment during purification is essential to the integrity of the final preparation and has a great impact on the phase separation propensity and activity of Rubisco. It is recommended that buffers contain 1-5 mM DTT at all times. Otherwise, Rubisco is forgiving and does not require addition of protease inhibitors or special treatment throughout purification. DTT must be added fresh to prevent its degradation.
Protocol
Lysing material
Note 1
This section of the protocol is very variable dependent on the nature of the material to be processed. It is recommended the final lysate is passed through a 0.2 μm filter prior to the next step but extraction techniques should be optimised on a material basis.
- Lyse material in 1x Rubisco lysis buffer (50 mM Tris-HCl, 100 mM NaCl, 0.5 mM EDTA, 5 mM DTT at pH 8.0).
- For green algae (Chlorella/Chlamydomonas), cell disruption by sonication or ‘French press (35 kPSI)’ has worked well in our hands.
- ~1 g of biomass (wet weight) can be efficiently lysed in ~7.5 mL of buffer, though excess lysis buffer is preferred, if practical.
- For plant and seaweed material, lysis using a household blender has worked very well in our hands.
- In this case, excess buffer almost always yields better lysis.
- For green algae (Chlorella/Chlamydomonas), cell disruption by sonication or ‘French press (35 kPSI)’ has worked well in our hands.
- Centrifuge lysate at ~50,000 g for 30 minutes at 4 °C.
- Depending on the efficiency of lysis, this step may need to be repeated. Alternatively, a lower speed (e.g. 5000 g, 30 minutes) spin could be used priort to this step, to remove large fragments and incompletely lysed cells.
- Filter supernatant through 0.2 µm membrane.
- At this point, the solution should be green, but free of large thylakoid fragments. Smaller thylakoid fragments will be removed in the following steps.
- If it is the first time performing an extraction on the specified material, here is a good place to take a sample for a gel to estimate your lysis efficiency.
Ammonium sulphate precipitation
Note 2
We have found ammonium sulphate precipitation very useful for removing smaller thylakoid fragments and crucially, contaminating chlorophyll-binding proteins (e.g. LHCBs), that will bind anion exchange media and reduce the efficiency and purity of subsequent purification steps. The precipitation is completed in two steps; one to remove unwanted large proteins and thylakoid proteins and a second to precipitate the remaining soluble proteins (including Rubisco). In short; discard the first pellet and keep the second!
- Meausure the volume of the lysate from the previous step in a measuring cylinder and dilute if necessary (e.g. if the solution is very dark/viscous).
- Ammonium sulphate precipitation in larger volumes is much more controllable and in turn appears to be more efficient. Dilution may also be required if the lysate is very dark (i.e. contains a large amount of pigment-binding proteins).
- Whilst the lysate is stirring, add ammonium sulphate powder to a final concentration of 190 mg mL-1 and stir for 1 hour at 4 °C.
- Centrifuge at ~50,000 g for 10 minutes at 4 °C.
At this stage, dark green pellets should be observed, which contain thylakoid material and unwanted large proteins. The supernatant should be largely free of green pigment, and have a yellow-ish colouration.

- Decant the supernatant and, whilst stirring, add ammonium sulphate powder to a final concentration of 390 mg mL-1 (a further 200 mg mL-1 to previous step) and stir for 1 hour at 4 °C.
- Centrifuge at ~50,000 g for 10 minutes at 4 °C.
- Very light green/brown pellets should be observed and contain most of the Rubisco.
- Resuspend the pelleted fraction in IEX buffer (A) (50 mM Tris-HCl, 50 mM NaCl, 1 mM DTT at pH 8.0).
- As a rough guide, we find the pellet from Chlorella can be resuspended to ~70% of the input mass (i.e. the pellet from 10 g of biomass can be resuspended in 7 mL of buffer)
- To check if the protein is sufficiently solubilised, centrifuge a small volume at ~20,000 g and check for a pellet (bad). Further dilute the suspension if pellets are still observed.
- Try to avoid the formation of bubbles during the resuspension.
Dialysis and lipid/hydrophobic protein removal
Note 3
Given the high concentration of salt used during the ammonium sulphate precipitation, it is important to dialyse the final suspension to remove contaminating salt in the final pellet. We have also anecdotally observed that the overnight dialysis performed here allows for the cohesion and subsequent removal of any remaining contaminating lipids.
- Dialyse the resuspended pelleted fraction against IEX Buffer (A) (50 mM Tris-HCl, 50 mM NaCl, 1 mM DTT at pH 8.0) overnight at 4 °C.
- Ensure that the volume of the dialysate is >100 the volume of the sample being dialysed.
- We use 14 kDa cut-off dialysis tubing for this step.
- Remove lipids and associated hydrophobic protein by centrifugation at ~90,000 g for 30 minutes at 4 °C.
- We have found this step to drastically improve the resolution of the subsequent IEX and avoids the contamination of the resin with lipids.
Anion exchange
Note 4
We use a 30 mL column self-packed with SOURCE 15Q resin for the anion exchange, and have found that lower quality resins are not sufficient to purify Rubisco to high purity in a single step.
- Pass the dialysed Rubisco fraction over the column equilibrated with IEX buffer (A).
- Wash the column back to ~0 mAU with IEX buffer (A) (~5 CVs).
- Step elute the majority of contaminating proteins with 15% IEX elution buffer (B) (to yield a concentration of 150 mM NaCl, 50 mM Tris-HCl, 1 mM DTT, pH 8.0)
- Elute the bound Rubisco with a linear gradient to 50% IEX elution buffer (B) (to yield 50 mM Tris-HCl, 0.5 M NaCl, 1 mM DTT at pH 8.0) over ~1 hour.
- Rubisco should elute between 225 mM and 300 mM NaCl:
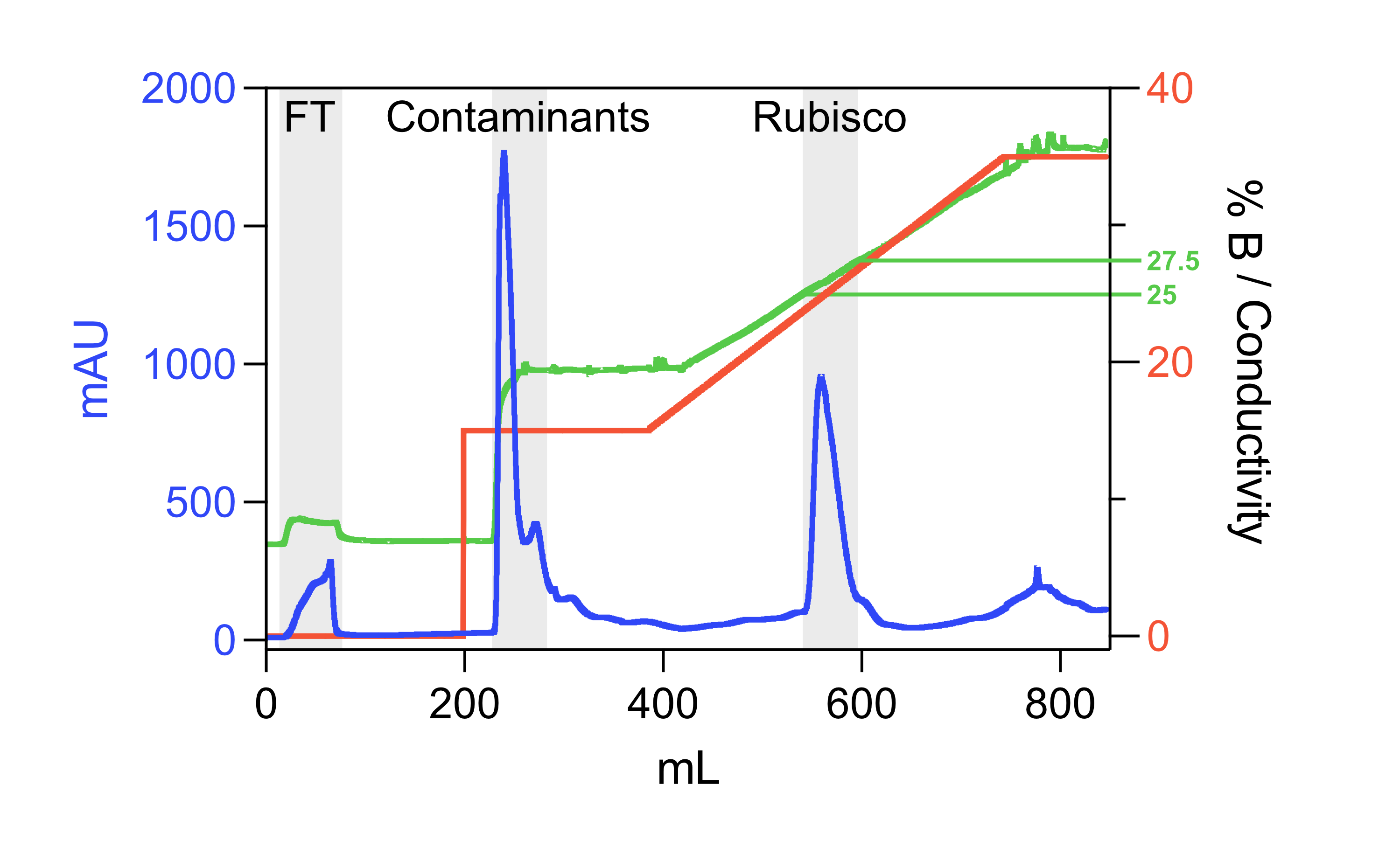
- Pool Rubisco fractions and concentrate (using e.g.</a>).
Size exclusion chromatography
Note 5
Size exclusion chromatography (SEC) should only be used as a final ‘polishing’ step. If your fractions still contain significant impurities, pigments, or nucleic acids, performing SEC will not yield significant improvements in purity.
- Equilibrate column with SEC buffer (50 mM Tris-HCl, 50 mM NaCl, 5% glycerol (w/v), 1 mM DTT at pH 8.0) and load Rubisco sample.
- We routinely use columns from the Superdex 200 preparative grade (pg) or increase families - typically 16/600 pg or 10/300 increase.
- The Rubisco holoenzyme elutes from S200 columns at ~47% of the column volume (i.e. 150 mL from 26/600 and 56 mL from 16/600.
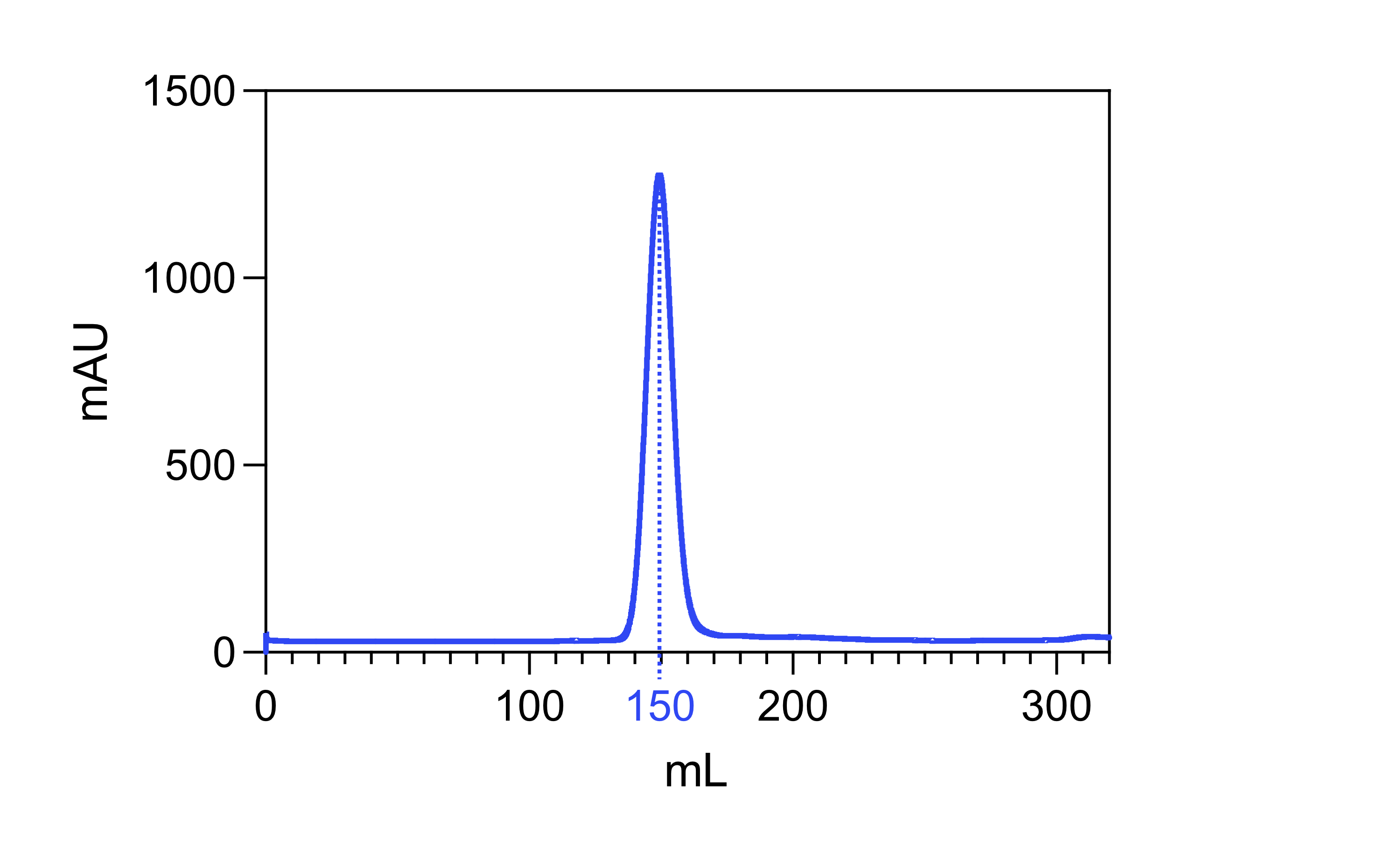
- Pool and concentrate the Rubisco fractions.
- The fraction can be flash-frozen in liquid nitrogen and stored at -70 °C in SEC buffer.
N.B.</br> Without reducing agents, we have observed the small subunits of the holoenzyme to fall off, leaving an L8 octamer and free small subunits in solution. This can be hard to detect without a known ‘good’ sample run adjacent on a native PAGE gel.
